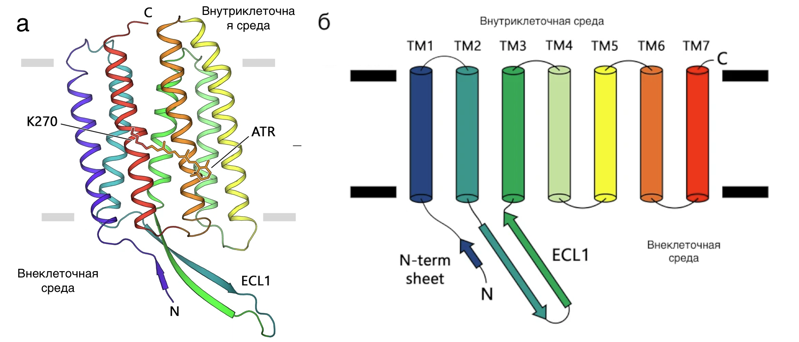
a) the structure of the studied rhodopsin, b) the scheme of the connection of protein helices.
Scientists from the Center for Research on Molecular Mechanisms of Aging and Age-related Diseases of the Moscow Institute of Physics and Technology, together with colleagues from Germany and the United States, for the first time obtained and studied the structure of a light-activated proton pump from mushrooms. It has been compared to known protein structures with the same function. It turned out that the protein has a common ancestor with the proton pump of microbes without membranes inside. However, it differs from the proteins of bacteria, the cells of which have membranes but no nucleus. The results obtained are important for further research on proteins in human cells. The work was published in the journal Communications Biology.
Living organisms are divided into three main groups (domains) according to the presence of membrane-limited areas in the composition of cells. Archaea (first domain) do not contain any membrane parts in cells. Bacteria are more complex (second domain) – organelles surrounded by membranes are present in their cells. Eukaryotes (third domain), in turn, contain both membrane organelles and a nucleus within cells. The latter group includes the kingdoms of animals (which include humans), fungi, plants, and some unicellular organisms. Evolutionary relationships between representatives of different groups of living things are studied by studying and comparing genomes and proteins.
Earlier comparisons of the genomes of many animals showed that light-sensitive proteins in cells perform extremely different functions and are present in all kingdoms, as well as in many large viruses. The most common family of light-sensitive proteins are rhodopsins. Rhodopsins of the first type consist of seven spirals piercing the cell membrane (Figure 1). Proteins use light energy for active and passive transport of ions across the cell membrane, triggering signaling reactions and activating enzymes in cells. Light-sensitive proteins are used in optogenetics, a technique for studying nerve impulses that can be excited with light. They are considered common light-harvesting proteins on Earth and the main light-capturing proteins in the oceans. Given the ubiquitous distribution of rhodopsins and their important ecological role, there is no doubt that these proteins played a significant role in the evolution of life on Earth. In the early stages of evolution, many organisms could use rhodopsins as one of the additional sources of energy, which gave them an evolutionary advantage. A wide variety of proton pumps, as well as their antiquity, make it possible to study global evolutionary processes based on the history of changes in these proteins. However, eukaryotic rhodopsins have been studied less extensively, since their isolation and crystallization are much more difficult.
The authors of the study used their recently developed expression methods (in the LEXSY system) to isolate type 1 rhodopsin from a unicellular fungus. Scientists have crystallized and obtained the structure of the protein. It turned out that the structure of rhodopsin from the fungus is very similar to the structure of the archaeal proton pump. The only significant difference was found in the intracellular part of the protein: the ECL1 loop is much longer than that of archaea rhodopsin (see Figure 1). The authors studied the function of this loop and found that it binds to the intracellular end, thereby increasing the stability of the protein. To determine the evolutionary relationship between rhodopsins, scientists compared the known structures and sequences of proteins. It turned out that the sequences and structures of eukaryotic rhodopsins are extremely similar to those of archaeal proteins. Apparently, rhodopsin genes undergo extensive horizontal transfer between organisms, which complicates the search for a common ancestor. However, the high structural similarity of the proton pumps of archaea and eukaryotes, along with their functional similarity, provides convincing evidence of the archaeal origin of eukaryotic proton rhodopsins and, most likely, all other eukaryotic rhodopsins.
“We obtained the first high-resolution crystal structure of a photosensitive proton pump from the organism of a fungus and elucidated the functional role of its N-terminal region. The studied rhodopsin was expressed in the LEXSY system, then we crystallized it. This means that the LEXSY expression system can be a powerful tool for obtaining eukaryotic membrane proteins for structural studies. We also compared the sequence and structure of the resulting rhodopsin with photosensitive proton pumps from different kingdoms. The analysis showed that eukaryotic and archaeal rhodopsins have a deep structural similarity, which confirms the hypothesis of the archaeal origin of rhodopsins found in the eukaryotic genome. The results obtained are important both for understanding the evolution of animals and for further research on eukaryotic rhodopsins, ”explains Dmitry Zabelsky, an employee of the Laboratory of Chemistry and Lipid Physics at the Center for Research on Molecular Mechanisms of Aging and Age-related Diseases, Moscow Institute of Physics and Technology, PhD student at the Phystech School of Physics and Research. Landau MIPT.
Read also:
Archaeologists have found two rare treasures in Staraya Ryazan and Suzdal Opolye
Chemists have proposed micro-generation of hydrogen from beer cans
An early Christian temple with a Byzantine cross and numerous burials was found near Chersonesos
Scientists have found a way to determine the homeland of frogs by their mucus
Zebrafish showed how to overcome chronic stress

